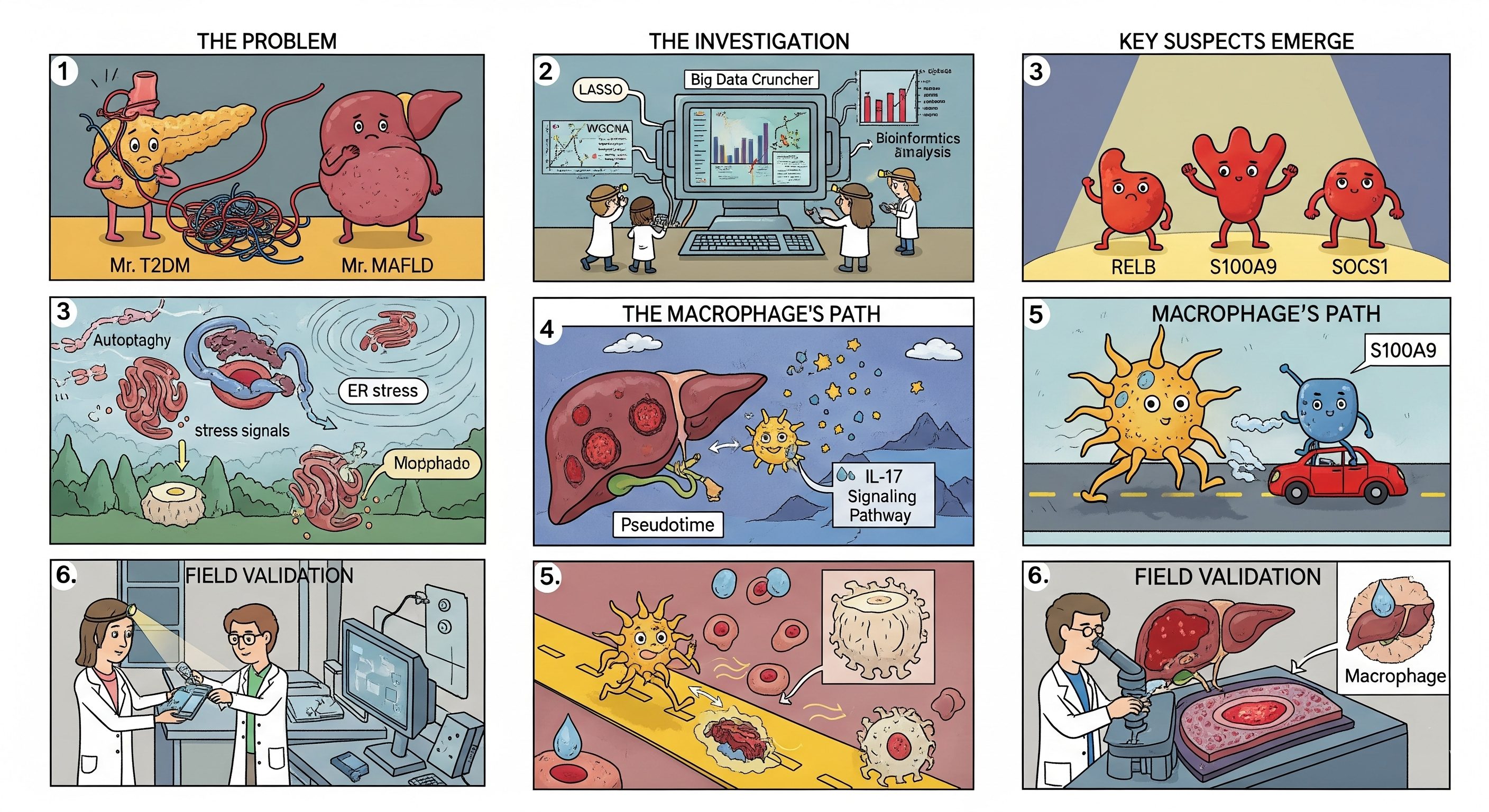
The metabolic collision between type 2 diabetes mellitus (T2DM) and metabolic dysfunction-associated fatty liver disease (MAFLD) fuels cardiometabolic risk, yet the molecular bridges that connect pancreatic, hepatic, and immune dysfunction remain elusive. This Scientific Reports study maps those bridges by merging bulk transcriptomes, single-cell atlases, and immunophenotyping to spotlight autophagy and endoplasmic reticulum stress pathways that could be druggable in both conditions.
Multi-Omic Integration Across Tissues
The authors started with four GEO datasets covering human liver and pancreatic tissues from T2DM and MAFLD cohorts. Differential expression intersected the two diseases, revealing 161 shared genes. Weighted gene co-expression network analysis (WGCNA), protein-protein interaction networks, and LASSO-based feature selection distilled the list to a quartet of hub genes—RELB, S100A9, SOCS1, and CDKN1A—that sit at the crossroads of autophagy and ER stress.
"Combining multi-cohort integration with single-cell validation allowed us to pinpoint autophagy-ER stress regulators that co-activate across endocrine and hepatic tissues," Wang et al. noted.
Immune Microenvironment Under the Microscope
Study Workflow at a Glance
Shared DEG Discovery
Intersected T2DM and MAFLD bulk RNA-seq datasets to extract common differentially expressed genes enriched for stress pathways.
Network Prioritisation
Applied WGCNA, PPI networks, and machine learning to prioritise RELB, S100A9, SOCS1, and CDKN1A as core regulators.
Immune Profiling
Used CIBERSORT to trace macrophage and neutrophil signatures tied to hub gene expression and metabolic stress.
Single-Cell Validation
Integrated scRNA-seq maps to assign hub gene expression to hepatocyte, Kupffer cell, and islet subpopulations.
What the Hub Genes Reveal
RELB and SOCS1 traced inflammatory signalling cascades, S100A9 highlighted neutrophil-driven damage, and CDKN1A reflected stress-induced cell-cycle arrest. Together they captured immune infiltration patterns that paralleled insulin resistance and steatosis severity. Multivariate models integrating the four genes outperformed traditional metabolic markers in classifying patients across external cohorts.
Bench Validation in Rat Models
Moving beyond in silico analyses, the team induced T2DM with MAFLD in Wistar rats. Histology, qRT-PCR, and immunohistochemistry confirmed that the four-gene signature jumps alongside steatosis, inflammation, and ER stress hallmarks. The animal data strengthen the translational relevance of targeting autophagy-ER stress crosstalk to blunt combined disease progression.
Discussion Topics in This Episode
- Connecting metabolic comorbidities: Why overlapping stress pathways make joint T2DM-MAFLD therapeutics plausible.
- Decoding immune infiltration: What CIBERSORT and scRNA-seq say about macrophages, neutrophils, and stellate cells.
- Biomarker translation: How four-gene panels could augment current risk scores for metabolic liver disease.
- Preclinical validation: Lessons from rat models that mirror human multi-organ metabolic stress.
- Drug discovery angles: Opportunities to modulate RELB-NF-κB signalling or S100A9-driven inflammation.
Key Takeaways
- Integrative transcriptomics converged on four stress-response genes that jointly stratify risk across T2DM and MAFLD.
- Immune infiltration patterns tied to RELB, S100A9, SOCS1, and CDKN1A suggest that inflammatory modulation could rebalance metabolic stress.
- Rat validation reinforces the potential of autophagy and ER stress pathways as shared therapeutic targets for intertwined metabolic diseases.
Resources and Further Reading
License Attribution
This podcast episode discusses research from: Wang, Q. et al. "Integrated bulk and single-cell transcriptomics identify RELB, S100A9, and SOCS1 as key autophagy-endoplasmic reticulum stress genes linking T2DM with MAFLD" Scientific Reports (2025). Licensed under CC BY 4.0.
Research Paper
Integrated bulk and single-cell transcriptomics identify RELB, S100A9, and SOCS1 as key autophagy-endoplasmic reticulum stress genes linking T2DM with MAFLD
Qin Wang et al. • Scientific Reports • 2025
Access the full text via the publisher link above under the CC BY 4.0 license.
Further Reading
- Recent consensus updates on metabolic dysfunction-associated steatotic liver disease and its overlap with T2DM.
- Reviews on autophagy-ER stress coupling in endocrine and hepatic tissues.
- Guides to applying CIBERSORT and single-cell integration in metabolic disease research.