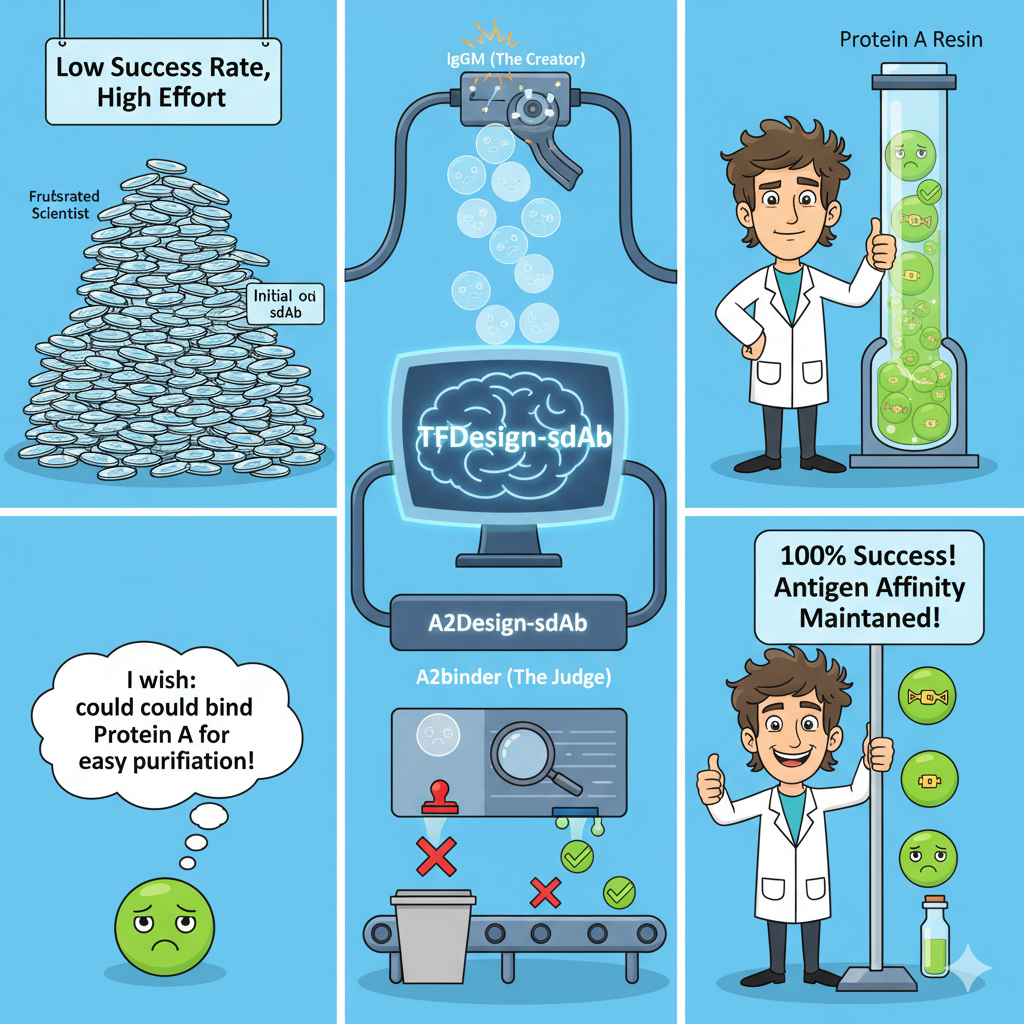
This week we explore how the TFDesign-sdAb pipeline takes single-domain antibodies that refuse to bind Protein A and teaches them new tricks. By pairing a diffusion generator with a structure-informed ranker, the team at Fudan University and Tencent AI Labs delivered nanobodies that can be purified on industry-standard resins while still latching onto their therapeutic targets.
Engineering Framework Regions with Diffusion Models
Traditional antibody engineering focuses on complementarity-determining regions, leaving framework residues untouched. TFDesign-sdAb flips that script by directing IgGM to co-design framework loops that contact Protein A domain D. The model operates in two training phases, first learning structure recovery and then sequence redesign, enabling precise edits across human VH scaffolds and camelid VHHs.
"Our diffusion-driven framework customization confers Protein A affinity while maintaining the original antigen paratope," Kong et al. report.
Ranking Candidates with Limited Experimental Data
Study Workflow at a Glance
Generative Design
IgGM samples 1000 sequence-structure variants per nanobody by conditioning on Protein A epitopes and framework contact residues.
Affinity Ranking
Fine-tuned A2binder filters candidates using a blend of antibody language models and convolutional interaction heads, despite limited sdAb training data.
Wet-Lab Validation
Top variants expressed in E. coli bind Protein A at tens of nanomolar KD values and can be eluted from commercial resin with efficiencies over 50%.
Structural Confirmation
1.49–3.57 Å crystal structures verify that engineered framework loops mirror the human VH Protein A binding geometry predicted in silico.
Maintaining Antigen Recognition
Despite introducing up to three framework mutations, redesigned nanobodies retained binding to their cognate antigens—CEACAM5, CD16A, TNFα, and the oncofetal antigen 5T4. ELISA dose-response curves showed EC50 values nearly identical to wild type, highlighting how targeted framework edits can add manufacturability traits without eroding therapeutic potency.
Why This Matters for Biologics Manufacturing
- Protein A capture is the gold standard for monoclonal antibody purification; extending it to sdAbs slashes downstream costs and removes immunogenic affinity tags.
- Framework-aware generative design enables additive engineering—augmenting antibodies with new functions while maintaining native antigen binding.
- High-resolution structures confirm that AI-selected residues recapitulate hydrogen-bond and salt-bridge networks essential for Protein A engagement.
Discussion Topics in This Episode
- Data efficiency: How fine-tuning with 30 in-house binding measurements lifted A2binder out of the zero-shot regime.
- Framework loops as design space: Why residues 72–76 and 90–92 emerged as hotspots for Protein A contact.
- Manufacturability metrics: Comparing chromatographic recovery and purity between Ni-NTA and Protein A workflows.
- Structural validation: Lessons from crystallographic overlays between native and AI-edited nanobodies.
- Next steps: Extending TFDesign-sdAb to engineer pharmacokinetics or effector functions beyond purification.
Resources and Further Reading
License Attribution
This episode discusses research from: Kong, Y. et al. "A synergistic generative-ranking framework for tailored design of therapeutic single-domain antibodies" Cell Discovery (2025). Licensed under CC BY 4.0.
Research Paper
A synergistic generative-ranking framework for tailored design of therapeutic single-domain antibodies
Yu Kong et al. • Cell Discovery • 2025
View on Publisher →Related Topics
- Diffusion models for antibody sequence and structure co-design.
- Framework engineering strategies for nanobody manufacturability.
- AI-assisted purification process development in biologics.