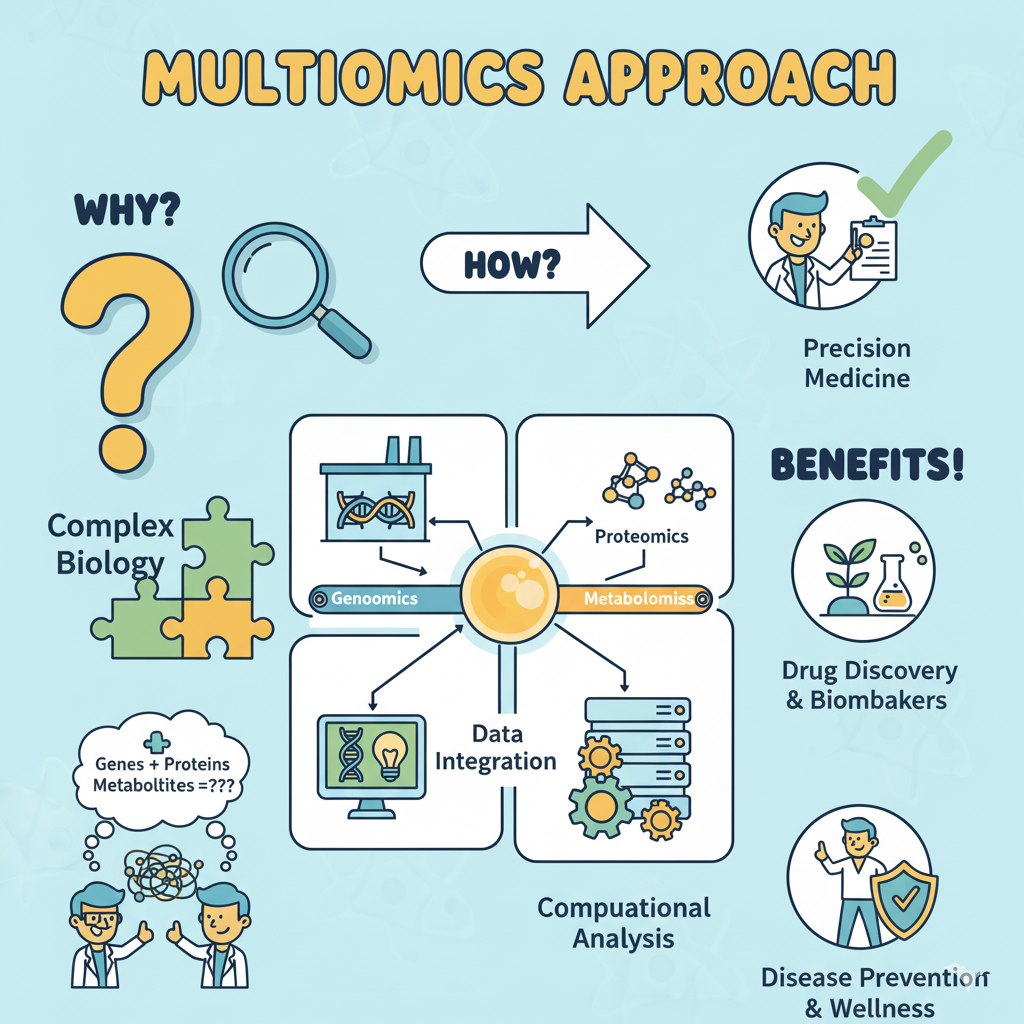
The last two decades gifted us unprecedented visibility into the genome, transcriptome, proteome, metabolome, and epigenome. Yet each domain alone delivers only a vignette. Biology operates as an interconnected system in which DNA variants ripple through RNA, proteins, and metabolites before manifesting as phenotypes. Multiomics acknowledges that reality. It integrates diverse molecular measurements to reveal how changes cascade across scales. For translational teams, it is the difference between reading isolated chapters and grasping the full narrative of disease.
What Do We Mean by Multiomics?
Multiomics refers to the coordinated generation, integration, and interpretation of multiple “omics” layers from the same biological system. In practice, that might mean pairing whole-genome sequencing with single-cell RNA-seq, bulk proteomics, phosphoproteomics, and targeted metabolomics for a patient cohort. Depending on the question, teams can add spatial transcriptomics, chromatin accessibility (ATAC-seq), lipidomics, or microbiome sequencing to round out the view.
The integrative power comes from both experimental design and computational strategy. Samples are collected using harmonized protocols, processed with consistent metadata standards, and analyzed within interoperable pipelines. Downstream, statistical modeling, machine learning, and knowledge graphs connect molecular nodes across datasets. When done well, multiomics does not just concatenate features; it contextualizes them.
Core omic layers most teams combine:
- • Genomics captures inherited and somatic variation that frame cellular potential.
- • Transcriptomics quantifies how genes are expressed across cell types, states, and conditions.
- • Proteomics and post-translational modifications reveal functional machinery and signaling activity.
- • Metabolomics and lipidomics expose pathway flux, energetic status, and phenotypic readouts.
- • Epigenomics and chromatin accessibility show how regulatory programs respond to internal or environmental cues.
Why Organizations Are Betting on Multiomics Now
Historically, cost, throughput, and analytical maturity limited multiomic studies to flagship consortia. That barrier is collapsing. Sequencing prices continue to fall, single-cell platforms are scaling, and data commons create interoperable backbones. Meanwhile, AI-driven analytics turn high-dimensional matrices into actionable signals. As a result, multiomics moved from an experimental luxury to a competitive necessity across biopharma, diagnostics, agriculture, and digital health.
Richer Disease Taxonomies
Integrated omics uncovers molecular subtypes that transcend traditional histology. Lung cancer, for example, fractures into genomic, immune, and metabolic clusters with distinct prognoses and therapeutic targets.
Predictive Biomarker Panels
Multi-feature models built from genomics plus proteomics routinely outperform single-omic signatures in predicting response to immunotherapy, guiding companion diagnostics, and stratifying clinical trials.
Systems-Level Drug Discovery
Drug hunters map upstream mutations to downstream pathway disruptions, uncovering combination targets and predicting resistance mechanisms before they emerge in the clinic.
Beyond human health, multiomics plays a pivotal role in microbiome engineering, crop optimization, environmental monitoring, and industrial biotech. Wherever complex phenotypes emerge from layered biology, integrative omics delivers leverage.
Five Tangible Benefits of Multiomic Programs
1. Earlier Signal Detection
Multiomics increases sensitivity to subtle perturbations. A genomic variant might look benign until transcriptomics reveals allele-specific expression, proteomics shows disrupted phosphorylation, and metabolomics detects disturbed glycolysis. Together, these hints flag risk years before symptoms manifest, enabling proactive interventions.
2. Mechanistic Confidence
Integrating causal chains clarifies why biomarkers shift. Instead of correlating a gene with disease, teams trace from mutation to transcript to protein function to metabolic flux. This mechanistic linking satisfies regulators, supports publication in high-impact journals, and accelerates translational decisions.
3. Personalized Therapeutic Design
Multiomic data enables patient-specific digital twins. Clinicians can simulate how a tumor’s mutational burden, immune infiltration, and metabolic dependencies will respond to combinatorial therapies, informing bespoke treatment plans.
4. Portfolio De-risking
For biopharma and diagnostics companies, multiomics reduces late-stage failure by catching liabilities earlier. Off-target effects, toxic pathway activations, and resistance trajectories surface before expensive Phase III trials or commercial launches.
5. Data Gravity for AI
Advanced foundation models thrive on multi-modal data. By constructing harmonized multiomic repositories today, organizations train proprietary AI systems that deliver defensible competitive advantage tomorrow.
Case Spotlight: Multiomics in Oncology
Precision oncology showcases multiomics at scale. The Pan-Cancer Analysis of Whole Genomes (PCAWG) and The Cancer Genome Atlas (TCGA) projects established foundational catalogs of mutations, expression programs, and epigenetic landscapes. Building atop those datasets, modern consortia now overlay proteomic signatures, immune repertoires, and single-cell atlases to decode tumor ecosystems.
"Tumors that appear identical under the microscope can diverge dramatically once you examine their multiomic profiles. That divergence matters when selecting immunotherapies, kinase inhibitors, or cell therapies."BioinfoXpert Oncology Practice Lead
Clinical labs are translating these insights into assays: combined DNA/RNA panels highlight actionable fusions; proteomic phospho-signatures predict kinase inhibitor response; metabolomic fingerprints flag metabolic vulnerabilities. Regulators increasingly accept integrated evidence packages, provided the data provenance and analytical validation are robust.
Building a High-Performing Multiomics Program
Successful multiomic initiatives demand orchestration across wet lab workflows, data operations, and analytics. At BioinfoXpert, we advise clients to stage their build-out across five pillars:
- Strategic Use-Case Framing: Anchor projects around questions that require cross-layer resolution such as mechanism of action clarification, resistance prediction, or biomarker discovery.
- Sample & Metadata Governance: Implement standardized collection kits, batch controls, and FAIR (Findable, Accessible, Interoperable, Reusable) metadata schemas to maintain comparability.
- Modular Lab Workflows: Use automation-friendly SOPs for nucleic acids, proteins, and metabolites. Cloud-connected LIMS track lineage and QC across modalities.
- Interoperable Data Platforms: Adopt storage formats (Parquet, AnnData, mzML) and ontologies (OBI, SNOMED, HGNC) that simplify cross-omic joins.
- Analytics and Interpretation: Pair statistical learning with knowledge graphs and domain expertise. Visualization dashboards should let bench scientists and clinicians interrogate findings together.
Operational Tip
Start with a pilot cohort (n = 30–50) to refine protocols, then scale. Use matched reference controls and replicate samples to calibrate integration methods before embarking on thousand-sample campaigns.
Overcoming Common Challenges
Despite the benefits, multiomics introduces complexity. Teams often hit the same hurdles like data volume, noise, and interpretability. Anticipating these issues keeps programs on track.
Data Deluge
Integrate tiered storage, data compression, and object stores to manage petabyte-scale matrices while keeping hot data accessible for analysis.
Batch & Platform Effects
Leverage mutual nearest neighbors, Harmony, or Liger to correct batch effects across omic layers, and include technical replicates to monitor drift.
Biological Interpretation
Couple AI-driven network inference with curated pathway databases and expert review cycles to avoid overfitting and ensure biological plausibility.
Ethical data stewardship also matters. Multiomic datasets carry sensitive genomic and clinical metadata. Programs must respect patient privacy, informed consent, and jurisdictional data residency rules. Differential privacy, federated learning, and synthetic data approaches can preserve utility while protecting individuals.
ROI: Measuring the Value of Multiomics
Executives often ask how to quantify multiomics ROI. The answer spans tangible financial metrics and strategic gains:
- Pipeline Efficiency: Track reduction in late-stage attrition, faster go/no-go decisions, and shorter cycle times from target ID to lead optimization.
- Diagnostic Accuracy: Measure improvements in sensitivity/specificity of assay panels and associated reimbursement approvals.
- Partnering & IP: Multiomic datasets underpin differentiated intellectual property and attract co-development deals.
- Talent Magnetism: High-quality data ecosystems help recruit and retain computational biologists, AI scientists, and physician-researchers.
For many organizations, the most compelling driver is future readiness. Multiomic infrastructures future-proof teams against emerging modalities (e.g., proteogenomics, single-cell spatial assays) and allow rapid pivots when new technologies appear.
The Role of Partners and Platforms
Few organizations have full-stack multiomics capabilities on day one. Strategic partnerships fill gaps, whether in assay development, computational pipelines, or domain interpretation. BioinfoXpert deploys modular services including pilot design, data engineering, AI modeling, and knowledge translation that align with client maturity. The goal is not perpetual outsourcing but capability uplift.
Ready to Connect Your Omics Data?
Our multiomics architects help research, clinical, and biotech teams design interoperable programs that deliver actionable insights in months, not years.
Start a Multiomics Blueprint →References & Further Reading
Want to go deeper? These publications and resources highlight the evidence base for multiomic integration across disease areas and analytical frameworks.
- Hasin, Y., Seldin, M. & Lusis, A. Multi-omics approaches to disease. Genome Biology 18, 83 (2017). https://doi.org/10.1186/s13059-017-1215-1
- Karczewski, K. J. & Snyder, M. P. Integrative omics for health and disease. Nature Reviews Genetics 19, 299–310 (2018). https://doi.org/10.1038/nrg.2018.4
- Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). https://doi.org/10.1016/j.cell.2019.05.031
- Liu, Y. et al. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nature Biotechnology 37, 314–322 (2019). https://doi.org/10.1038/s41587-019-0037-y
- Gillette, M. A. et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 182, 200–225 (2020). https://doi.org/10.1016/j.cell.2020.06.013
- Zhang, B. et al. Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 (2014). https://doi.org/10.1038/nature13438
- Dou, Y. et al. Proteogenomic characterization of endometrial carcinoma. Cell 180, 729–748.e26 (2020). https://doi.org/10.1016/j.cell.2020.01.026
- The Cancer Genome Atlas Research Network & Clinical Proteomic Tumor Analysis Consortium. Integrated proteogenomic characterization of high-grade serous ovarian cancer. Cell 166, 755–765 (2016). https://doi.org/10.1016/j.cell.2016.05.069